What is protein cross-linking?
Protein crosslinking usually refers to the joining of two amino acids in a protein by a covalent bond other than a peptide bond. Protein crosslinking includes both intermolecular and intramolecular crosslinking, which are distinguished by whether the amino acids attached at the ends of the crosslink are from the same protein. The connected parts can be divided into exogenous protein crosslinks and endogenous protein crosslinks according to their sources.
Exogenous Crosslinking
A chemical reagent is added to a protein solution to link two functional groups of amino acid side chains in a protein or protein complex. This type of crosslinking is a tool to study the spatial structure and interactions of proteins, without affecting the structure and function of the protein itself.
The formation of exogenous crosslinks preserves information on protein topology, conformational state and protein-protein interactions (PPIs) in the form of cross-linked peptides.
The amino acid residues that can form crosslinks are usually relatively close together in space, and this distance depends on the arm length of the crosslinker, so that the tertiary structure of the protein can be inferred from the information of the crosslinking sites. Protein interactions are usually transient and they can be immobilized using crosslinkers so that they will always be retained during subsequent processing.
Exogenous crosslinks are formed by the binding of amino acids by an applied chemical crosslinker. Therefore, when using mass spectrometry for the identification of exogenous crosslinks, various functional groups can be added specifically to the chemical crosslinker to make the fragmentation with a specific pattern, simplifying the identification of crosslinks. Moreover, the chemical properties of exogenous crosslinks are relatively stable, and the types of peptides forming crosslinks are relatively homogeneous.
Endogenous Crosslinking
Unlike exogenous crosslinks, the formation of endogenous crosslinks relies on small molecules or chemical bonds present in the cell to link amino acid residues together. Endogenous crosslinking occurs naturally within proteins and is a specific post-translational modification of proteins. Its formation includes both enzymatic and non-enzymatic catalysis and has important implications for the structure and function of proteins.
Compared to exogenous crosslinks that are chemically dependent, endogenous crosslinks are not chemically stable enough and are prone to heterodimerization and fracture when using mass spectrometry for data acquisition. Therefore, the study of endogenous crosslinks using mass spectrometry usually requires the investigation of the fragmentation pattern before an accurate identification of endogenous crosslinks can be achieved.
In a single protein, multiple endogenous crosslinks can exist. It has been reported that protein crosslinks, including isopeptide bonds and disulfide bonds, are found in 30% of Bacillus subtilis spore shell proteins.
The endogenous crosslinks are both reversible and irreversible. They play a key role in maintaining protein structural stability, redox regulation, etc.
Endogenous protein crosslinks have been identified, including disulfide bonds, isopeptide bonds, tryptophan crosslinks, tyrosine crosslinks, formaldehyde crosslinks, methylglyoxal (MG) crosslinks, and NOS bridges.
Cross-linking Mass Spectrometry for Protein Cross-linking Detection
Chemical cross-linking combined with mass spectrometry uses a chemical cross-linker to treat a protein sample, linking two amino acids that are close enough in space to react with the cross-linker in a covalent bond, and then analyzing the cross-linked products using mass spectrometry-based proteomics methods.
Cross-linking mass spectrometry is mainly used for the study of protein structure and protein interactions. The immobilization of the cross-linker allows the retention of many loosely structured and easily dissociated complexes.
Workflow of cross-linked mass spectrometry
The whole process can be basically divided into 7 stages.
Stage 1: The protein sample to be analyzed is separated from the cell lysate or tissue by biochemical separation or affinity selection.
Stage 2: A chemical cross-linking reaction is performed by adding a chemical cross-linking agent to the protein sample to form protein cross-links. Endogenous cross-linking is naturally present in proteins, so the second stage in the above step can be skipped.
Stage 3: The sample obtained in the previous stage is degraded into peptide fragments by the action of protease in preparation for subsequent peptide sequencing.
Phase 4: The obtained peptides pass through the elution of liquid chromatography and enter the electrospray chamber of the mass spectrometer. Under the action of strong electric field, the eluted peptides can be charged, which enables the mass spectrometer to analyze the peptides.
Phase 5: The mass spectrometer analyzes the charged eluted peptides by mass spectrometry to obtain the first level spectrum (MS1 spectrum).
Stage 6: The computer generates a priority list of these peptides and then performs a tandem mass spectrometry analysis to obtain a secondary spectrum (MS2 spectrum).
Stage 7: Qualitative and quantitative analysis of peptides and proteins is achieved by analyzing the spectra generated by the above process.
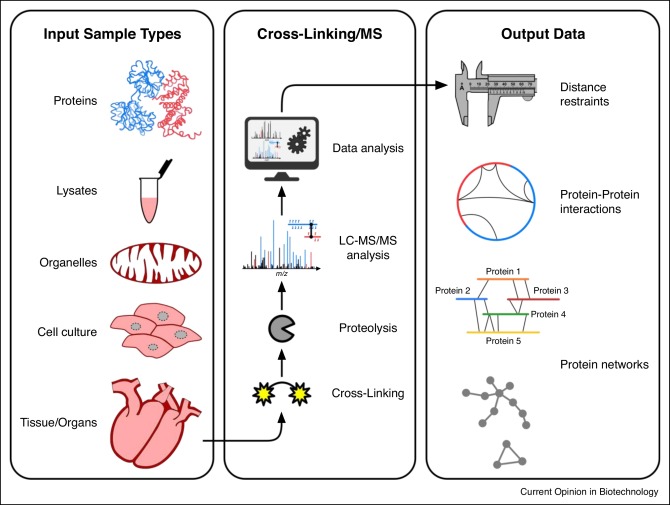
XL-MS workflows (Iacobucci et al., 2020)
Creative Proteomics offers protein cross-linking services as well as cross-linking mass spectrometry analysis services. If you are interested, please feel free to contact us.
Reference
- Iacobucci, C., Götze, M., & Sinz, A. (2020). Cross-linking/mass spectrometry to get a closer view on protein interaction networks. Current opinion in biotechnology, 63, 48-53.
Read More: